Tohban Guide to Self Calibration and Imaging for EOVSA
This tutorial describes the step-by-step procedure to download EOVSA IDB data, obtain and calibrate the measurement sets (.ms), and, transfer them to the inti server for self-calibration and further analysis.
Pre-requisites
Accounts on Pipeline and Inti or Baozi servers. Sijie and Bin can help with the account set-up for pipeline, inti and boazi.
How to connect to pipeline
Windows: On a windows machine Mobaxterm can be used to connect to Pipeline. The process to add rsa keys to access gues@ovsa is slightly complicated. Please contact Dale for the set up.
Linux/Unix: On a linux/Mac, the guest account will be used for people to log in as Tohban. Rather than using a password, access will be tied to specific computers that you use. One needs to generate a public + private ssh key pair using ssh-keygen or equivalent program that will result in a public key (plain text) file called id_rsa.pub, created in your ~/.ssh folder, the contents of which can be emailed to Dale or Owen to be added under "authorized keys". The ssh commands can then be used directly through the terminal:
ssh -L 5902:helios.solar.pvt:20000 guest@ovsa.njit.edu ### will not ask for a password for your computer's authorized key ### ssh -XY your_username@pipeline ### add your username and enter password to your pipeline account, when prompted ###
Alternatively, to avoid typing the longer commands, proxy commands can be added to the ~/.ssh/config file:
vim ~/.ssh/config ### press 'i' to insert content and esc +':wq' to write and quit ###
Host *
ForwardAgent yes
Host *
ForwardX11 yes
Host ovsa
HostName ovsa.njit.edu
User guest
Host pipeline
Hostname pipeline.solar.pvt
User your_username ### Insert your pipeline username here ###
ProxyCommand ssh -W %h:%p guest@ovsa.njit.edu
Upon saving the ~/.ssh/config file and restarting the terminal, you can connect to pipeline directly using:
ssh -XY pipeline
How to connect to the inti server
NOTE: To connect to inti you must have a NJIT UCID. The arcs have granted access to inti for all SRG members. If you encounter any issue with the login. Please let arcs folks (arcs@njit.edu) know.
Windows: As mentioned above, on a windows machine Mobaxterm can be used to connect to inti. With your NJIT VPN connected, connect to one of the afsconnect servers (for example, afsaccess3.njit.edu) using your UCID and password. Next, ssh into inti.
ssh -XY UCID@afsaccess3.njit.edu ###Insert UCID password if prompted### ssh -XY UCID@inti.hpcnet.campus.njit.edu ###Insert UCID/inti username here###
Proxy commands for inti can also be added to the C:\Program Files\Git\etc\ssh\ssh_config file:
Host inti Hostname inti.hpcnet.campus.njit.edu User UCID ###Insert UCID/inti username here###
Linux/Unix: On a linux/Mac the ssh command can be used:
ssh -XY UCID@inti.hpcnet.campus.njit.edu ### insert UCID here and enter your NJIT UCID password, when prompted ###
Proxy commands for inti can also be added to the ~/.ssh/config (on local machine) file for convenience:
vim ~/.ssh/config ### press 'i' to insert content and esc +':wq' to write and quit ###
Host inti
Hostname inti.hpcnet.campus.njit.edu
User UCID ### insert NJIT UCID here ###
after the proxy is set up, connect to inti by simply typing:
ssh -XY inti ### enter your NJIT UCID password when prompted ###
NOTE:When logging in for the first time, please add the following lines (demarcated by #) in your ~/.bashrc (on the inti server) file in order to use CASA on inti.
#### setting start #### if [ $HOSTNAME == "baozi.hpcnet.campus.njit.edu" ]; then source /srg/.setenv_baozi fi if [ $HOSTNAME == "inti.hpcnet.campus.njit.edu" ]; then source /inti/.setenv_inti fi if [ $HOSTNAME == "guko.resource.campus.njit.edu" ]; then source /data/data/.setenv_guko fi #### setting end ####
For sufficient disk space to analyze EOVSA data on Inti, use your dedicated directory YOURDIRECTORY at the location given below. If you do not have a directory, please ask Sijie to set one up.
cd /inti/data/users/YOURDIRECTORY
Both CASA 5 and 6 are available on Inti.
Enter the bash environment on inti, and load the desired casa environment.
To load CASA 5:
>> bash >> loadcasa5 >> suncasa
To load CASA 6:
>> bash >> loadcasa6 >> ipython
Downloading data and Preliminary Calibration
Step 1: Downloading raw (IDB) data on Pipeline machine
The initial downloading of data and applying refcal and phasecal to the .ms files is performed on the pipeline since the complete EOVSA SQL database is stored here for now.
If you are not using mobaxterm, directly SSH to Pipeline through your Linux or Mac computer (as shown above) and start tcsh to run CASA.
The following commands (run within CASA) can be used to import a single IDB file if the given time-range is less than 10 mins.
from astropy.time import Time
import os
trange = Time(['2017-08-21 20:15:00', '2017-08-21 20:25:00']) ###Change accordingly###
#### (Optional) change output path, default current directory "./" #######
outpath = './msdata/' ###Change accordingly###
if not os.path.exists(outpath):
os.makedirs(outpath)
msfiles = importeovsa(idbfiles=trange, ncpu=1, visprefix=outpath)
NOTE: The above step currently gives an error with importeovsa. This is because the data location was changed and the code doesn't know where to look for the IDB files. Sijie is looking into the problem. We will update the page when it is fixed. For now, please use the method below for all time ranges.
OR (for a time range longer than 10 minutes)
from suncasa.tasks import task_calibeovsa as calibeovsa
from suncasa.tasks import task_importeovsa as timporteovsa
from split_cli import split_cli as split
import dump_tsys as dt
from util import Time
import numpy as np
import os
from glob import glob
from eovsapy import util
trange = Time(['2017-08-21 20:15:00', '2017-08-21 20:35:00']) ###Change accordingly###
idbdir = util.get_idbdir(trange[0])
info = dt.rd_fdb(trange[0])
for k, v in info.iteritems():
info[k] = info[k][~(info[k] == '')]
sidx = np.where(
np.logical_and(info['SOURCEID'] == 'Sun', info['PROJECTID'] == 'NormalObserving') & np.logical_and(
info['ST_TS'].astype(np.float) >= trange[0].lv,
info['ST_TS'].astype(np.float) <= trange[
1].lv))
filelist = info['FILE'][sidx]
outpath = './msdata/' ###Change accordingly###
if not os.path.exists(outpath):
os.makedirs(outpath)
inpath = idbdir + '{}/'.format(trange[0].datetime.strftime("%Y%m%d"))
ncpu = 1
msfiles = timporteovsa.importeovsa(idbfiles=[inpath + ll for ll in filelist], ncpu=ncpu, timebin="0s", width=1,
visprefix=outpath,
nocreatms=False, doconcat=False,
modelms="", doscaling=False, keep_nsclms=False, udb_corr=True)
If you come across errors with calibeovsa, add the following lines to your ~/.casa/init.py (within pipeline) file.
vim ~/.ssh/config ### press 'i' to insert content and esc +':wq' to write and quit ###
import sys
sys.path.append('/common/python')
sys.path.append('/common/python/packages/pipeline_casa')
execfile('/common/python/suncasa/tasks/mytasks.py')
Step 2: Concatenate multiple .ms files and Calibrate
Follow this step if there are more than one .ms files for the chosen time-range. If not, move to Step 3.
concatvis = os.path.basename(msfiles[0])[:11] + '_concat_cal.ms' ### Set the path/name for the concatenated files ###
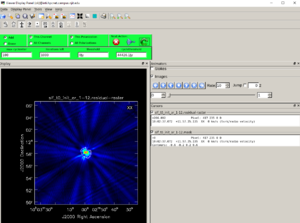
vis = calibeovsa.calibeovsa(msfiles, doconcat=True, concatvis=concatvis, caltype=['refpha','phacal'], doimage=True)
### If doimage=True, a quicklook image will be produced (by integrating over the entire time) as shown in Figure 1 ###
vis_str = str(' '.join(vis))
caled_vis=vis_str.replace('.ms','_cal.ms') ### Append '_cal' to the ms filename ###
split(vis=' '.join(vis),outputvis=caled_vis,datacolumn='corrected',timerange='',correlation='XX') ### Split the corrected column to the new 'caled' ms ###
Step 3: Calibration
For a single .ms file from Step 1, this will calibrate the input visibility, write out calibration tables under /data1/eovsa/caltable/, and apply the calibration.
vis = calibeovsa.calibeovsa('IDB20170821202020.ms', caltype=['refpha','phacal'], doimage=True) ###Change the vis filename accordingly###
vis_str = str(' '.join(vis))
caled_vis=vis_str.replace('.ms','_cal.ms') ### Append '_cal' to the ms filename ###
split(vis=' '.join(vis),outputvis=caled_vis,datacolumn='corrected',timerange='',correlation='XX') ### Split the corrected column to the new 'caled' ms ###
The created 'caled' ms data files (xxx_concat_cal.ms or xxx_cal.ms) need to be transferred from pipeline to inti server, which has all the casa tasks installed, in order to run the rest of the imaging steps.
Transferring data files between servers
To directly transfer your calibrated .ms data from the Pipeline to Inti:
1. Log into Inti using your username. ssh -X USERNAME@inti.hpcnet.campus.njit.edu Then create a tunnel into Pipeline from Inti. ssh -L 8888:pipeline.solar.pvt:22 guest@ovsa.njit.edu 2. Log into Inti again from a new terminal. Change to your working directory (where you need the data transferred) and give the following command: scp -v -C -r -P 8888 PipelineUSERNAME@localhost:PATH/TO/MSFILE ./
Advanced Calibration and Imaging
Step 4: Self-calibration

Follow the steps below to run the self-calibration of the imaging data and produce the calibrated images in .fits format. https://github.com/binchensun/casa-eovsa/blob/master/slfcal_example.py
bash
loadcasa5
suncasa
from suncasa.utils import helioimage2fits as hf
import os
import numpy as np
import pickle
from matplotlib import gridspec as gridspec
from sunpy import map as smap
from matplotlib import pyplot as plt
from split_cli import split_cli as split
import time
# =========== task handlers =============
dofullsun = 1 # initial full-sun imaging ###Change accordingly###
domasks=1 # get masks ###Change accordingly###
doslfcal=1 # main cycle of doing selfcalibration ###Change accordingly###
doapply=1 # apply the results ###Change accordingly###
doclean_slfcaled=1 # perform clean for self-calibrated data ###Change accordingly###
# ============ declaring the working directories ============
workdir = os.getcwd()+'/' #main working directory. Using current directory in this example
slfcaldir = workdir+'slfcal/' #place to put all selfcalibration products
imagedir = slfcaldir+'images/' #place to put all selfcalibration images
maskdir = slfcaldir+'masks/' #place to put clean masks
imagedir_slfcaled = slfcaldir+'images_slfcaled/' #place to put final self-calibrated images
caltbdir = slfcaldir+'caltbs/' # place to put calibration tables
# make these directories if they do not already exist
dirs = [workdir, slfcaldir, imagedir, maskdir, imagedir_slfcaled, caltbdir]
for d in dirs:
if not os.path.exists(d):
os.makedirs(d)
# ============ Split a short time for self-calibration ===========
# input visibility
ms_in = workdir + 'IDB20170821202020_cal.ms' ###Change the initial calibrated (through calibeovsa) vis accordingly###
# output, selfcaled, visibility
ms_slfcaled = workdir + os.path.basename(ms_in).replace('cal','slfcaled')
# intermediate small visibility for selfcalbration
# selected time range for generating self-calibration solutions
trange='2017/08/21/20:21:10~2017/08/21/20:21:30' ###Change accordingly###
slfcalms = slfcaldir+'slfcalms.XX.slfcal'
slfcaledms = slfcaldir+'slfcalms.XX.slfcaled'
if not os.path.exists(slfcalms):
split(vis=ms_in,outputvis=slfcalms,datacolumn='data',timerange=trange,correlation='XX')
# ============ Prior definitions for spectral windows, antennas, pixel numbers =========
spws=[str(s+1) for s in range(30)] ###spws=[str(s+1) for s in range(49)] # For post-2020 data
antennas='0~12&0~12'
npix=512
nround=3 #number of slfcal cycles
# =========== Step 1, doing a full-Sun image to find out phasecenter and appropriate field of view =========
if dofullsun:
#initial mfs clean to find out the image phase center
im_init='fullsun_init'
os.system('rm -rf '+im_init+'*')
tclean(vis=slfcalms,
antenna=antennas,
imagename=im_init,
spw='2~15',
specmode='mfs',
timerange=trange,
imsize=[npix],
cell=['5arcsec'],
niter=1000,
gain=0.05,
stokes='I',
restoringbeam=['30arcsec'],
interactive=False,
pbcor=True)
hf.imreg(vis=slfcalms,imagefile=im_init+'.image.pbcor',fitsfile=im_init+'.fits',
timerange=trange,usephacenter=False,verbose=True)
clnjunks = ['.flux', '.mask', '.model', '.psf', '.residual','.sumwt','.pb','.image'] ###Do not run the next 4 lines if needed to view and assess the subset of clean process images
for clnjunk in clnjunks:
if os.path.exists(im_init + clnjunk):
os.system('rm -rf '+im_init + clnjunk)
from sunpy import map as smap
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(6,6))
ax = fig.add_subplot(111)
eomap=smap.Map(im_init+'.fits')
#eomap.data=eomap.data.reshape((npix,npix))
eomap.plot_settings['cmap'] = plt.get_cmap('jet')
eomap.plot(axes = ax)
eomap.draw_limb()
plt.show()
viewer(im_init+'.image.pbcor')
# parameters specific to the event (found from step 1)
phasecenter='J2000 10h02m59 11d58m07' ###Change accordingly###
xran=[280,480] ###Change accordingly###
yran=[-50,150] ###Change accordingly###
# =========== Step 2 (optional), generate masks =========
# The outlines drawn for masks can be created by any of the icons with the letter 'R' in the Viewer window. The instructions for doing this can be found by hovering over those icons. See Figure 4.
# if skipped, will not use any masks
if domasks:
clearcal(slfcalms)
delmod(slfcalms)
antennas=antennas
pol='XX'
imgprefix=maskdir+'slf_t0'
# step 1: set up the clean masks
img_init=imgprefix+'_init_ar_'
os.system('rm -rf '+img_init+'*')
#spwrans_mask=['1~5','6~12','13~20','21~30']
spwrans_mask=['2~12']
#convert to a list of spws
spwrans_mask_list = [[str(i) for i in (np.arange(int(m.split('~')[0]),int(m.split('~')[1])))] for m in spwrans_mask] # Not used?
masks=[]
imnames=[]
for spwran in spwrans_mask:
imname=img_init+spwran.replace('~','-')
try:
tclean(vis=slfcalms,
antenna=antennas,
imagename=imname,
spw=spwran,
specmode='mfs',
timerange=trange,
imsize=[npix],
cell=['2arcsec'],
niter=1000,
gain=0.05,
stokes='XX',
restoringbeam=['20arcsec'],
phasecenter=phasecenter,
weighting='briggs',
robust=1.0,
interactive=True,
datacolumn='data',
pbcor=True,
savemodel='modelcolumn')
imnames.append(imname+'.image')
masks.append(imname+'.mask')
clnjunks = ['.flux', '.model', '.psf', '.residual'] #Do not run the next 4 lines if needed to view and assess the subset of clean process images
for clnjunk in clnjunks:
if os.path.exists(imname + clnjunk):
os.system('rm -rf '+ imname + clnjunk)
except:
print('error in cleaning spw: '+spwran)
pickle.dump(masks,open(slfcaldir+'masks.p','wb'))
# =========== Step 3, main step of selfcalibration =========
if doslfcal:
if os.path.exists(slfcaldir+'masks.p'):
masks=pickle.load(open(slfcaldir+'masks.p','rb'))
if not os.path.exists(slfcaldir+'masks.p'):
print 'masks do not exist. Use default mask'
masks=[]
os.system('rm -rf '+imagedir+'*')
os.system('rm -rf '+caltbdir+'*')
#first step: make a mock caltable for the entire database
print('Processing ' + trange)
slftbs=[]
calprefix=caltbdir+'slf'
imgprefix=imagedir+'slf'
tb.open(slfcalms+'/SPECTRAL_WINDOW')
reffreqs=tb.getcol('REF_FREQUENCY')
bdwds=tb.getcol('TOTAL_BANDWIDTH')
cfreqs=reffreqs+bdwds/2.
tb.close()
# starting beam size at 3.4 GHz in arcsec #Change accordingly
sbeam=40.
strtmp=[m.replace(':','') for m in trange.split('~')]
timestr='t'+strtmp[0]+'-'+strtmp[1]
refantenna='0'
# number of iterations for each round
niters=[100, 300, 500]
# roburst value for weighting the baselines
robusts=[1.0, 0.5, 0.0]
# apply calibration tables? Set to true for most cases
doapplycal=[1,1,1]
# modes for calibration, 'p' for phase-only, 'a' for amplitude only, 'ap' for both
calmodes=['p','p','a']
# setting uvranges for model image (optional, not used here)
uvranges=['','','']
for n in range(nround):
slfcal_tb_g= calprefix+'.G'+str(n)
fig = plt.figure(figsize=(8.4,7.)) # fig = plt.figure(figsize=(14, 7)) # For post-2020 data (50 spws)
gs = gridspec.GridSpec(5, 6) # gs = gridspec.GridSpec(5, 10) # For post-2020 data (50 spws)
for s,sp in enumerate(spws):
print 'processing spw: '+sp
cfreq=cfreqs[int(sp)]
# setting restoring beam size (not very useful for selfcal anyway, but just to see the results)
bm=max(sbeam*cfreqs[1]/cfreq, 6.)
slfcal_img = imgprefix+'.spw'+sp.zfill(2)+'.slfcal'+str(n)
# only the first round uses nearby spws for getting initial model
if n == 0:
spbg=max(int(sp)-2,1) # spbg=max(int(sp)-2,0) # For post-2020 data (50 spws)
sped=min(int(sp)+2,30) # sped=min(int(sp)+2,49) # For post-2020 data (50 spws)
spwran=str(spbg)+'~'+str(sped)
print('using spw {0:s} as model'.format(spwran))
if 'spwrans_mask' in vars():
for m, spwran_mask in enumerate(spwrans_mask):
if sp in spwran_mask:
mask = masks[m]
print('using mask {0:s}'.format(mask))
findmask = True
if not findmask:
print('mask not found. Do use any masks')
else:
spwran = sp
if 'spwrans_mask' in vars():
for m, spwran_mask in enumerate(spwrans_mask):
if sp in spwran_mask:
mask = masks[m]
print 'using mask {0:s}'.format(mask)
findmask = True
if not findmask:
print('mask not found. Do use any masks')
try:
tclean(vis=slfcalms,
antenna=antennas,
imagename=slfcal_img,
uvrange=uvranges[n],
spw=spwran,
specmode='mfs',
timerange=trange,
imsize=[npix],
cell=['2arcsec'],
niter=niters[n],
gain=0.05,
stokes='XX', #use pol XX image as the model
weighting='briggs',
robust=robusts[n],
phasecenter=phasecenter,
mask=mask,
restoringbeam=[str(bm)+'arcsec'],
pbcor=False,
interactive=False,
savemodel='modelcolumn')
if os.path.exists(slfcal_img+'.image'):
fitsfile=slfcal_img+'.fits'
hf.imreg(vis=slfcalms,imagefile=slfcal_img+'.image',fitsfile=fitsfile,
timerange=trange,usephacenter=False,toTb=True,verbose=False,overwrite=True)
clnjunks = ['.mask','.flux', '.model', '.psf', '.residual', '.image','.pb','.image.pbcor','.sumwt'] #Do not run the next 4 lines if needed to view and assess the subset of clean process images
for clnjunk in clnjunks:
if os.path.exists(slfcal_img + clnjunk):
os.system('rm -rf '+ slfcal_img + clnjunk)
ax = fig.add_subplot(gs[s])
eomap=smap.Map(fitsfile)
eomap.plot_settings['cmap'] = plt.get_cmap('jet')
eomap.plot(axes = ax)
eomap.draw_limb()
#eomap.draw_grid()
ax.set_title(' ')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
ax.set_xlim(xran)
ax.set_ylim(yran)
plt.pause(0.5) # Allows viewing of each image as it is plotted.
os.system('rm -f '+ fitsfile)
except:
print 'error in cleaning spw: '+sp
print 'using nearby spws for initial model'
sp_e=int(sp)+2
sp_i=int(sp)-2
if sp_i < 1: # if sp < 0: # For post-2020 data (50 spws)
sp_i = 1 # sp_i = 0 # For post-2020 data (50 spws)
if sp_e > 30: # if sp_e > 49: # For post-2020 data (50 spws)
sp_e = 30 # sp_e = 49 # For post-2020 data (50 spws)
sp_=str(sp_i)+'~'+str(sp_e)
try:
tget(tclean)
spw=sp_
print('using spw {0:s} as model'.format(sp_))
tclean()
except:
print 'still not successful. abort...'
break
gaincal(vis=slfcalms, refant=refantenna,antenna=antennas,caltable=slfcal_tb_g,spw=sp, uvrange='',\
gaintable=[],selectdata=True,timerange=trange,solint='inf',gaintype='G',calmode=calmodes[n],\
combine='',minblperant=4,minsnr=2,append=True)
if not os.path.exists(slfcal_tb_g):
print 'No solution found in spw: '+sp
figname=imagedir+'slf_t0_n{:d}.png'.format(n)
plt.subplots_adjust(left=0, bottom=0, right=1, top=1, wspace=0, hspace=0)
plt.savefig(figname)
time.sleep(10)
plt.close()
if os.path.exists(slfcal_tb_g):
slftbs.append(slfcal_tb_g)
slftb=[slfcal_tb_g]
os.chdir(slfcaldir)
if calmodes[n] == 'p':
plotcal(caltable=slfcal_tb_g,antenna='1~12',xaxis='freq',yaxis='phase',\
subplot=431,plotrange=[-1,-1,-180,180],iteration='antenna',figfile=slfcal_tb_g+'.png',showgui=False)
if calmodes[n] == 'a':
plotcal(caltable=slfcal_tb_g,antenna='1~12',xaxis='freq',yaxis='amp',\
subplot=431,plotrange=[-1,-1,0,2.],iteration='antenna',figfile=slfcal_tb_g+'.png',showgui=False)
os.chdir(workdir)
plt.pause(0.5) # Allows viewing of the plot
if doapplycal[n]:
clearcal(slfcalms)
delmod(slfcalms)
applycal(vis=slfcalms,gaintable=slftb,spw=','.join(spws),selectdata=True,\
antenna=antennas,interp='nearest',flagbackup=False,applymode='calonly',calwt=False)
if n < nround-1:
prompt=raw_input('Continuing to selfcal?')
#prompt='y'
if prompt.lower() == 'n':
if os.path.exists(slfcaledms):
os.system('rm -rf '+slfcaledms)
split(slfcalms,slfcaledms,datacolumn='corrected')
print 'Final calibrated ms is {0:s}'.format(slfcaledms)
break
if prompt.lower() == 'y':
slfcalms_=slfcalms+str(n)
if os.path.exists(slfcalms_):
os.system('rm -rf '+slfcalms_)
split(slfcalms,slfcalms_,datacolumn='corrected')
slfcalms=slfcalms_
else:
if os.path.exists(slfcaledms):
os.system('rm -rf '+slfcaledms)
split(slfcalms,slfcaledms,datacolumn='corrected')
print 'Final calibrated ms is {0:s}'.format(slfcaledms)
# =========== Step 4: Apply self-calibration tables =========
if doapply:
import glob
os.chdir(workdir)
clearcal(ms_in)
clearcal(slfcalms)
applycal(vis=slfcalms,gaintable=slftbs,spw=','.join(spws),selectdata=True,\
antenna=antennas,interp='linear',flagbackup=False,applymode='calonly',calwt=False)
applycal(vis=ms_in,gaintable=slftbs,spw=','.join(spws),selectdata=True,\
antenna=antennas,interp='linear',flagbackup=False,applymode='calonly',calwt=False)
if os.path.exists(ms_slfcaled):
os.system('rm -rf '+ms_slfcaled)
split(ms_in, ms_slfcaled,datacolumn='corrected')
# =========== Step 5: Generate final self-calibrated images (optional) =========
if doclean_slfcaled:
import glob
pol='XX'
print('Processing ' + trange)
img_final=imagedir_slfcaled+'/slf_final_{0}_t0'.format(pol)
vis = ms_slfcaled
spws=[str(s+1) for s in range(30)]
tb.open(vis+'/SPECTRAL_WINDOW')
reffreqs=tb.getcol('REF_FREQUENCY')
bdwds=tb.getcol('TOTAL_BANDWIDTH')
cfreqs=reffreqs+bdwds/2.
tb.close()
sbeam=30.
from matplotlib import gridspec as gridspec
from sunpy import map as smap
from matplotlib import pyplot as plt
fitsfiles=[]
for s,sp in enumerate(spws):
cfreq=cfreqs[int(sp)]
bm=max(sbeam*cfreqs[1]/cfreq,4.)
imname=img_final+'_s'+sp.zfill(2)
fitsfile=imname+'.fits'
if not os.path.exists(fitsfile):
print 'cleaning spw {0:s} with beam size {1:.1f}"'.format(sp,bm)
try:
tclean(vis=vis,
antenna=antennas,
imagename=imname,
spw=sp,
specmode='mfs',
timerange=trange,
imsize=[npix],
cell=['1arcsec'],
niter=1000,
gain=0.05,
stokes=pol,
weighting='briggs',
robust=2.0,
restoringbeam=[str(bm)+'arcsec'],
phasecenter=phasecenter,
mask='',
pbcor=True,
interactive=False)
except:
print 'cleaning spw '+sp+' unsuccessful. Proceed to next spw'
continue
if os.path.exists(imname+'.image.pbcor'):
imn = imname+'.image.pbcor'
hf.imreg(vis=vis,imagefile=imn,fitsfile=fitsfile,
timerange=trange,usephacenter=False,toTb=True,verbose=False)
fitsfiles.append(fitsfile)
junks=['.flux','.model','.psf','.residual','.mask','.image','.pb','.image.pbcor','.sumwt'] #Do not run the next 4 lines if needed to view and assess the subset of clean process images
for junk in junks:
if os.path.exists(imname+junk):
os.system('rm -rf '+imname+junk)
else:
print('fits file '+fitsfile+' already exists, skip clean...')
fitsfiles.append(fitsfile)
fig = plt.figure(figsize=(8.4,7.)) # fig = plt.figure(figsize=(14, 7)) # For post-2020 data (50 spws)
gs = gridspec.GridSpec(5, 6) # gs = gridspec.GridSpec(5, 10) # For post-2020 data (50 spws)
for s,sp in enumerate(spws):
cfreq=cfreqs[int(sp)]
ax = fig.add_subplot(gs[s])
eomap=smap.Map(fitsfiles[s])
eomap.plot_settings['cmap'] = plt.get_cmap('jet')
eomap.plot(axes = ax)
eomap.draw_limb()
ax.set_title(' ')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
ax.set_xlim(xran)
ax.set_ylim(yran)
plt.text(0.98,0.85,'{0:.1f} GHz'.format(cfreq/1e9),transform=ax.transAxes,ha='right',color='w',fontweight='bold')
plt.subplots_adjust(left=0, bottom=0, right=1, top=1, wspace=0, hspace=0)
plt.show()
The .fits files of the self calibrated images at each frequency for the given time are saved at /slfcal/images_slfcaled in your working directory.
Step 5: Quick-look imaging
For spectral imaging analysis of the event, follow this tutorial using the self-calibrated data obtained from the previous step or use this link.
from suncasa.utils import qlookplot as ql ## (Optional) Supply the npz file of the dynamic spectrum from previous step.
## If not provided, the program will generate a new one from the visibility.
## set the time interval
from suncasa import dspec as ds
import time
visibility_data = 'IDB20170821202020_slfcaled.ms'
specfile = visibility_data + '.dspec.npz'
d = ds.Dspec(visibility_data, bl='4&9', specfile=specfile)
timerange = '19:02:00~19:02:10' ## select frequency range from 2.5 GHz to 3.5 GHz
spw = '2~5' ## select stokes XX
stokes = 'XX' ## turn off AIA image plotting, default is True
plotaia = False
xycen = [375, 45] ## image center for clean in solar X-Y in arcsec
cell=['2.0arcsec'] ## pixel size
imsize=[128] ## x and y image size in pixels.
fov = [100,100] ## field of view of the zoomed-in panels in unit of arcsec
spw = ['{}'.format(s) for s in range(1,31)]
clevels = [0.5, 1.0] ## contour levels to fill in between.
calpha=0.35 ## now tune down the alpha
restoringbeam=['6arcsec']
ql.qlookplot(vis=msfile, specfile=specfile, timerange=timerange, spw=spw, stokes=stokes, \
restoringbeam=restoringbeam,imsize=imsize,cell=cell, \
xycen=xycen,fov=fov,clevels=clevels,calpha=calpha)
Page Last Updated: 07/07/2022